Experiment¶
Experiment is the playground to prepare training and testing data, and search the optimized estimator with HyperModel. Use experiment with the following steps:
- Prepare datasets
- training data(X_train, y_train): required
- evaluation data(X_eval, y_eval): optional, will be split from training data if not provided
- testing data(X_test only): optional, the sample of the prediction data, is used to optimize the experiment
- Create HyperModel instance with your selected implementation
- The PlainModel is provided as a plain implementation in this project.
- More implementations can be found from DeepTables, HyperGBM, HyperKeras, etc.
- Create experiment instance with the prepared datasets and HyperModel instance and other arguments required by your selected experiment implementation ( GeneralExperiment , CompeteExperiment , or your customized implementation).
- Call experiment’s .run() to search the optimized estimator.
GeneralExperiment¶
GeneralExperiment is the basic implementation of experiment. Example code:
from sklearn.model_selection import train_test_split
from hypernets.experiment import GeneralExperiment
from hypernets.searchers import make_searcher
from hypernets.tabular.datasets import dsutils
def create_hyper_model(reward_metric='auc', optimize_direction='max'):
from hypernets.examples.plain_model import PlainModel, PlainSearchSpace
search_space = PlainSearchSpace()
searcher = make_searcher('random', search_space_fn=search_space, optimize_direction=optimize_direction)
hyper_model = PlainModel(searcher=searcher, reward_metric=reward_metric, callbacks=[])
return hyper_model
def general_experiment_with_heart_disease():
hyper_model = create_hyper_model()
X = dsutils.load_heart_disease_uci()
y = X.pop('target')
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3)
experiment = GeneralExperiment(hyper_model, X_train, y_train, eval_size=0.3)
estimator = experiment.run(max_trials=5)
score = estimator.evaluate(X_test, y_test, metrics=['auc', 'accuracy', 'f1', 'recall', 'precision'])
print('evaluate score:', score)
if __name__ == '__main__':
general_experiment_with_heart_disease()
CompeteExperiment¶
CompeteExperiment is an implementation of experiment with many advanced features for tabular data, such as data cleaning, feature generation, feature selection, semi-supervised machine learning, two-stages searching etc.
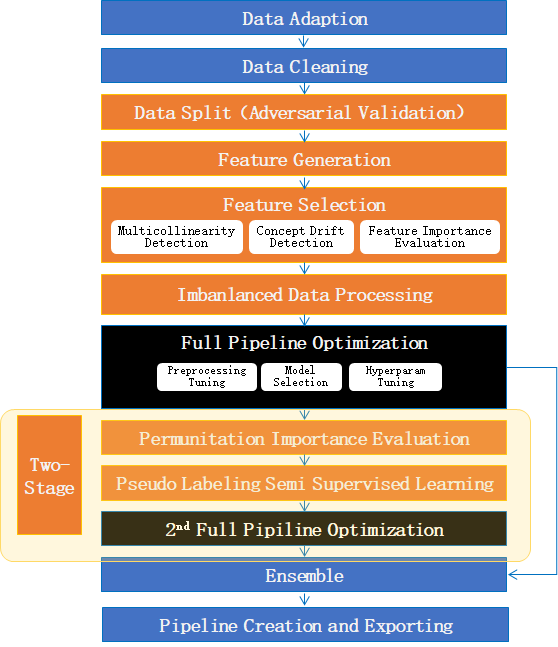
Quick start¶
Basically, use CompeteExperiment with default settings just like GeneralExperiment.
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
from hypernets.experiment import CompeteExperiment
from hypernets.tabular.datasets import dsutils
from hypernets.tabular.metrics import calc_score
def create_hyper_model(reward_metric='auc', optimize_direction='max'):
from hypernets.core.callbacks import SummaryCallback
from hypernets.examples.plain_model import PlainModel, PlainSearchSpace
from hypernets.searchers import make_searcher
from hypernets.tabular.sklearn_ex import MultiLabelEncoder
search_space = PlainSearchSpace(enable_dt=True, enable_lr=True, enable_nn=False)
searcher = make_searcher('random', search_space_fn=search_space, optimize_direction=optimize_direction)
hyper_model = PlainModel(searcher=searcher, reward_metric=reward_metric, callbacks=[SummaryCallback()],
transformer=MultiLabelEncoder)
return hyper_model
def experiment_with_bank_data(row_count=3000):
X = dsutils.load_bank()
if row_count is not None:
X = X.head(row_count)
X['y'] = LabelEncoder().fit_transform(X['y'])
y = X.pop('y')
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=9527)
experiment = CompeteExperiment(create_hyper_model(), X_train, y_train, max_trials=10)
estimator = experiment.run()
preds = estimator.predict(X_test)
proba = estimator.predict_proba(X_test)
score = calc_score(y_test, preds, proba, metrics=['auc', 'accuracy', 'f1', 'recall', 'precision'])
print('evaluate score:', score)
assert score
if __name__ == '__main__':
experiment_with_bank_data()
Set the Number of Search Trials¶
One can set the max search trial number by adjusting max_trials.
The following codes set the max trial times as 300:
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model, max_trials=300, ...)
Use Cross Validation¶
Users can apply cross validation in the experiment by manually setting parameter cv. Setting cv as ‘False’ will lead the experiment to avoid using cross validation and apply train_test_split instead. On the other hand, when cv is True, the experiment will use cross validation where the number of folds can be adjusted through the parameter num_folds. The default value of num_folds is 3.
Example code when cv=True:
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model, cv=True, num_folds=5, ...)
Evaluation dataset¶
When cv=False, training model will require evaluating its performance additionally on evaluation dataset. This can be done by setting X_eval and y_eval when creating CompeteExperiment. For example:
df = dsutils.load_blood()
X = df.copy()
y = X.pop(target)
X_train, X_eval, y_train, y_eval = train_test_split(X, y , test_size=0.3)
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model, X_train=X_train,y_train=y_train, X_eval=X_eval, y_eval=y_eval, ...)
If the X_eval or y_eval is None, the experiment object will split the X_train and y_train to get an evaluation dataset, whose size can be adjusted by setting eval_size:
df = dsutils.load_blood()
X = df.copy()
y = X.pop(target)
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model, X_train=X, y_train=y, eval_size=0.3, ...)
Set the Evaluation Criterion¶
The default evaluation criterion is accuracy for classification task is, and rmse for regression task. Other criterions can be set by reward_metric. For example:
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model, reward_metric='auc', ...)
Set the Early Stopping¶
One can set the early stopping strategy with settings of early_stopping_round, early_stopping_time_limit and early_stopping_reward.
The following code sets the max searching time as 3 hours:
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model, max_trials=300, early_stopping_time_limit=3600 * 3, ...)
Choose a Searcher¶
One can choose a specific searcher for the experiment by setting the parameter searcher.
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model, searcher='random', ...)
Furthermore, you can customize a new searcher object for experiment, for an example:
from hypernets.searchers import MCTSSearcher
my_searcher = MCTSSearcher(lambda: search_space_general(n_estimators=100),
max_node_space=20,
optimize_direction='max')
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model, searcher=my_searcher, ...)
Ensemble Models¶
CompeteExperiment automatically turns on the model ensemble function to get a better model when created. It will ensemble the best 20 models while the number for ensembling can be changed by setting ensemble_size as the following code, where ensemble_size=0 means to disable ensembling.
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model, ensemble_size=10, ...)
Data Adaption¶
This step supports Pandas/Cuml data types only, relevant parameters:
- data_adaption:(default True). Whether to enable data adaption.
- data_adaption_memory_limit:(default 0.05). If float, should be between 0.0 and 1.0 and represent the proportion of the system free memory. If int, represents the absolute byte number of memory.
- data_adaption_min_cols:(default 0.3. If float, should be between 0.0 and 1.0 and represent the proportion of the original dataframe column number. If int, represents the absolute column number.
- data_adaption_target:(default None),Where to run the next steps. ‘cuml’ or ‘cuda’, adapt training data into cuml datatypes and run next steps on nvidia GPU Devices. None, not change the training data types.
Data cleaning¶
CompeteExperiment performs data cleaning with DataCleaner in Hypernets. Note that this step can not be disabled but can be adjusted with DataCleaner in the following ways:
- nan_chars: value or list, (default None), replace some characters with np.nan
- correct_object_dtype: bool, (default True), whether correct the data types
- drop_constant_columns: bool, (default True), whether drop constant columns
- drop_duplicated_columns: bool, (default False), whether delete repeated columns
- drop_idness_columns: bool, (default True), whether drop id columns
- drop_label_nan_rows: bool, (default True), whether drop rows with target values np.nan
- replace_inf_values: (default np.nan), which values to replace np.nan with
- drop_columns: list, (default None), drop which columns
- reserve_columns: list, (default None), reserve which columns when performing data cleaning
- reduce_mem_usage: bool, (default False), whether try to reduce the memory usage
- int_convert_to: bool, (default ‘float’), transform int to other types,None for no transformation
If nan is represented by ‘\N’ in data,users can replace ‘\N’ back to np.nan when performing data cleaning as follows:
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model, data_cleaner_args={'nan_chars': r'\N'}, ...)
...
Feature generation¶
CompeteExperiment is capable of performing feature generation, which can be turned on by setting feature_generation=True when creating experiment with make_experiment. There are several options:
- feature_generation_continuous_cols:list (default None)), continuous feature, inferring automatically if set as None.
- feature_generation_categories_cols:list (default None)), categorical feature, need to be set explicitly, CompeteExperiment can not perform automatic inference for this one.
- feature_generation_datetime_cols:list (default None), datetime feature, inferring automatically if set as None.
- feature_generation_latlong_cols:list (default None), latitude and longtitude feature, inferring automatically if set as None.
- feature_generation_text_cols:list (default None), text feature, inferring automatically if set as None.
- feature_generation_trans_primitives:list (default None), transformations for feature generation, inferring automatically if set as None.
When feature_generation_trans_primitives=None, CompeteExperiment will automatically infer the types used for transforming based on the default features. Specifically, different transformations will be adopted for different types:
- continuous_cols: None, need to be set explicitly.
- categories_cols: cross_categorical.
- datetime_cols: month, week, day, hour, minute, second, weekday, is_weekend.
- latlong_cols: haversine, geohash
- text_cols:tfidf
An example code for enabling feature generation:
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model, feature_generation=True, ...)
Please see featuretools for more information.
Collinearity detection¶
There will often be some highly relevant features which are not informative but are more seen as noises. They are not very useful. On the contrary, the dataset will be affected by drifts of these features more heavily.
It is possible to handle these collinear features with CompeteExperiment. This can be simply enabled by setting collinearity_detection=True when creating experiment.
Example code for using collinearity detection
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model, collinearity_detection=True, ...)
...
Drift detection¶
Concept drift is one of the major challenge for machine learning. The model will often perform worse in practice due to the fact that the data distributions will change along with time. To handle this problem, CompeteExperiment adopts Adversarial Validation to detect whether there is any drifted features and drop them to maintain a good performance.
To enable drift detection, one needs to set drift_detection=True when creating experiment and provide X_test.
Relevant parameters:
- drift_detection_remove_shift_variable : bool, (default=True), whether to detect the stability of every column first.
- drift_detection_variable_shift_threshold : float, (default=0.7), stability socres higher than this value will be dropped.
- drift_detection_threshold : float, (default=0.7), detecting scores higher than this value will be dropped.
- drift_detection_remove_size : float, (default=0.1), ratio of columns to be dropped.
- drift_detection_min_features : int, (default=10), the minimal number of columns to be reserved.
- drift_detection_num_folds : int, (default=5), the number of folds for cross validation.
An code example:
from io import StringIO
import pandas as pd
from hypergbm import make_experiment
from hypernets.tabular.datasets import dsutils
test_data = """
Recency,Frequency,Monetary,Time
2,10,2500,64
4,5,1250,23
4,9,2250,46
4,5,1250,23
4,8,2000,40
2,12,3000,82
11,24,6000,64
2,7,1750,46
4,11,2750,61
1,7,1750,57
2,11,2750,79
2,3,750,16
4,5,1250,26
2,6,1500,41
"""
df = dsutils.load_blood()
X = df.copy()
y = X.pop(target)
test_df = pd.read_csv(StringIO(test_data))
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model, X_train=X, y_train=y, X_test=test_df,
drift_detection=True, ...)
...
Feature selection¶
CompeteExperiment evaluates the feature importance by training a pre-defined model. Then it chooses the most important ones among them to continue the model training.
To enable feature selection, one needs to set feature_selection=True when creating experiment. Relevant parameters:
- feature_selection_strategy:str, selection strategies(default threshold), can be chose from threshold, number and quantile.
- feature_selection_threshold:float, (default 0.1), selection threshold when the strategy is threshold, features with scores higher than this threshold will be selected.
- feature_selection_quantile:float, (default 0.2), selection threshold when the strategy is quantile, features with scores higher than this threshold will be selected.
- feature_selection_number:int or float, (default 0.8), selection numbers when the strategy is number.
An example code:
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model,
feature_selection=True,
feature_selection_strategy='quantile',
feature_selection_quantile=0.3,
...)
UnderSampling pre-search¶
Normally, hyperparameter optimization utilizes all training data. However, this will cost a huge amount of time for a large dataset. To alleviate this problem, one can perform a pre-search with only a part of data to try more model parameters in the same amount of time. Better parameters will then be used for training with the whole data to obtain the optimal parameters.
To enable feature selection, one needs to set down_sample_search=True when creating experiment. Relevant parameters:
- down_sample_search_size:int, float(0.0~1.0) or dict (default 0.1), number of examples used for pre-search.
- down_sample_search_time_limit:int, (default early_stopping_time_limit*0.33), time limit for pre-search.
- down_sample_search_max_trials:int, (default max_trials*3), max trail numbers for pre-search.
An example code:
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model,
down_sample_search=True,
down_sample_search_size=0.2,
...)
The second stage feature selection¶
CompeteExperiment supports continuing data processing with the trained model, which is officially called Two-stage search. There are two types of Two-stage processing supported by CompeteExperiment: Two-stage feature selection and pseudo label which will be covered in the rest of this section.
In CompeteExperiment, the second stage feature selection is to choose models with good performances in the first stage, and use permutation_importance to evaluate them to give better features.
To enable the second stage feature selection, one needs to set feature_reselection=True when creating experiment. Relevant parameters:
- feature_reselection_estimator_size:int, (default=10), the number of models to be used for evaluating the importances of feature (top n best models in the first stage).
- feature_reselection_strategy:str, selection strategy(default threshold), available selection strategies include threshold, number, quantile.
- feature_reselection_threshold:float, (default 1e-5), threshold when the selection strategy is threshold, importance scores higher than this values will be choosed.
- feature_reselection_quantile:float, (default 0.2), threshold when the selection strategy is quantile, importance scores higher than this values will be choosed.
- feature_reselection_number:int or float, (default 0.8), the number of features to be selected when the strategy is number.
An example code:
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model,
feature_reselection=True,
...)
Please refer to scikit-learn for more information about permutation_importance.
Pseudo label¶
Pseudo label is a kind of semi-supervised machine learning method. It will assign labels predicted by the model trained in the first stage to some examples in test data. Then examples with higher confidence values than a threshold will be added into the trainig set to train the model again.
To enable feature selection, one needs to set pseudo_labeling=True when creating experiment. Relevant parameters:
- pseudo_labeling_strategy:str, selection strategy(default threshold), available strategies include threshold, number and quantile.
- pseudo_labeling_proba_threshold:float(default 0.8), threshold when the selection strategy is threshold, confidence scores higher than this values will be chose.
- pseudo_labeling_proba_quantile:float(default 0.8), threshold when the selection strategy is quantile, importance scores higher than this values will be chose.
- pseudo_labeling_sample_number:float(0.0~1.0) or int (default 0.2), the number of top features to be selected when the strategy is number.
- pseudo_labeling_resplit:bool(default=False), whether split training and validation set after adding pseudo label examples. If set as False, all examples with pseudo labels will be added into training set to train the model. Otherwise, experiment will perform training set and validation set splitting for the new dataset with pseudo labels.
An example code:
from hypergbm import make_experiment
X_test=...
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model,
X_test=X_test,
pseudo_labeling=True,
...)
Note: Pseudo label is only valid for classification task.
Change the log level¶
The progress messages during training can be printed by setting log_level (str or int) to change the log level. Please refer to the logging package for more details. Besides, more thorough messages will show when verbose is set as 1.
The following codes sets the log level to ‘INFO’:
hyper_model = create_hyperModel()
experiment = CompeteExperiment(hyper_model, log_level='INFO', verbose=1, ...)
Export experiment report¶
If you want to export the experiment report in Excel format after training, you can set the argument as report_render= ‘excel’. The sample codes for displaying the experiment report are shown as follows:
from sklearn.model_selection import train_test_split
from hypernets.examples.plain_model import PlainModel, PlainSearchSpace
from hypernets.experiment import make_experiment
from hypernets.tabular.datasets import dsutils
df = dsutils.load_boston()
df_train, df_eval = train_test_split(df, test_size=0.2)
search_space=PlainSearchSpace(enable_lr=False, enable_nn=False, enable_dt=False, enable_dtr=True)
experiment = make_experiment(PlainModel, df_train,
target='target',
search_space=search_space,
report_render='excel')
estimator = experiment.run(max_trials=3)
print(estimator)
An excel report file named report.xlsx will be generated in the current directory. The report include the information of the engineering features, evaluation scores, resource usage and ensemble models, as well as the automatically generated plots. One example is shown below:
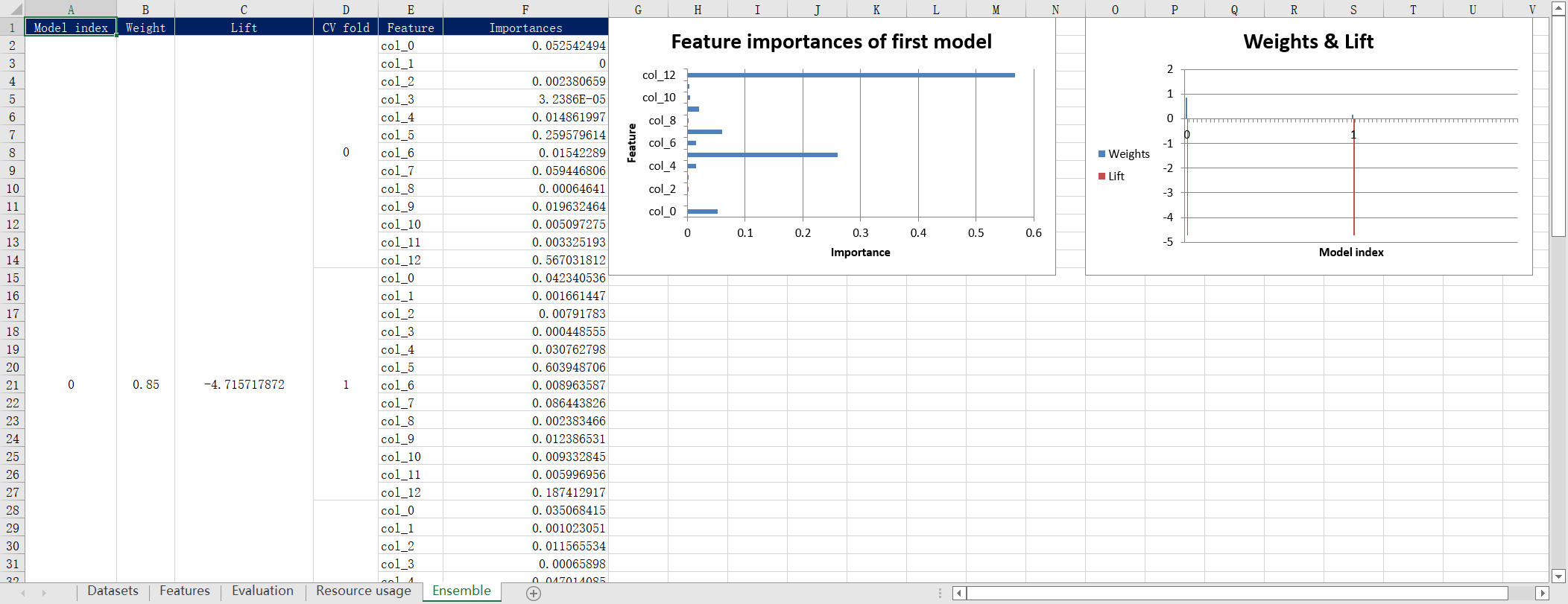
You can also change the file path of the output report by setting the argument report_render_options, here is the example code of how to set output report to /tmp/report.xlsx:
...
experiment = make_experiment(...,
report_render='excel',
report_render_options={'file_path': "/tmp/report.xlsx"})
...
Experiment visualization in jupyter notebook¶
Hypernets support two new features of experiment visualization in notebooks:
- visualization of the experiment configurations
- visualization of the dataset information
These features are optional tools and do not influence the experiment results.
To install the notebook visualization tool, use the command:
pip install hypernets[notebook]
If you have not installed the notebook widget, you can install it by command:
pip install hboard-widget
Here is an example of how to use these features in jupyter notebook :
- Import the required modules
from sklearn.model_selection import train_test_split
from hypernets.examples.plain_model import PlainModel, PlainSearchSpace
from hypernets.experiment import make_experiment
from hypernets.tabular.datasets import dsutils
- Create a hypernets experiment
df_train, df_eval = train_test_split(df, test_size=0.2)
search_space=PlainSearchSpace(enable_lr=False, enable_nn=False, enable_dt=False, enable_dtr=True)
experiment = make_experiment(PlainModel, df_train,
target='target',
search_space=search_space)
experiment
- The experiment configurations are automatically displayed, which provides a convenient reference for further modification.
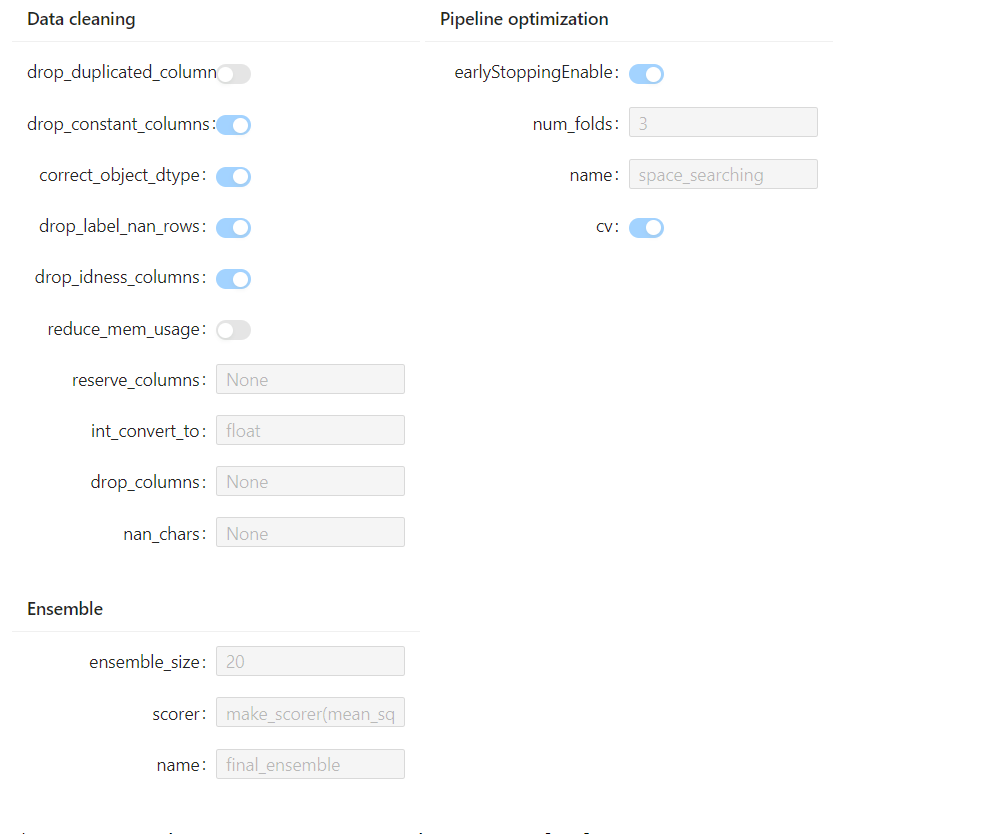
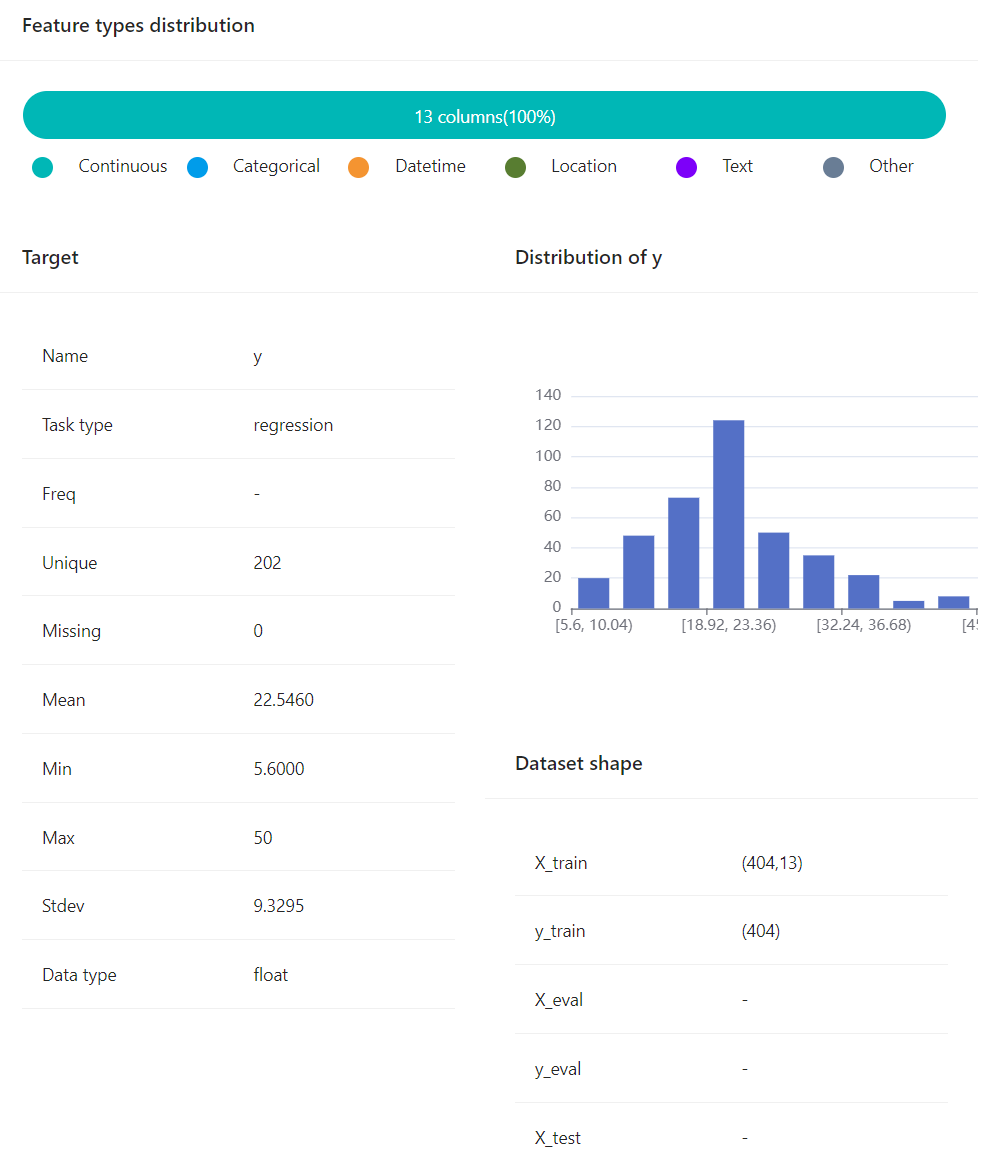
- The dataset information can also be easily visualized by command:
experiment.plot_dataset()
You can find the notebook example at hypernets_experiment_notebook_visualization.ipynb